A report from the RCDD diagnostic development group detailing the design of a molecular test for the species determination of Gram-negative bacteria has been published in Scientific Reports.
Gram-negative bacteria include many important human pathogens, and are known for their ability to develop extensive drug resistance. Dr Thomas Edwards, the lead author said ‘the determination of the species responsible for a Gram-negative bacterial infection is an important step for guiding patient management by tailoring antimicrobial therapy, and early identification is known to improve clinical outcomes. Current biochemical and culture based methods are time consuming, and molecular testing can reduce the time taken to provide these results’.
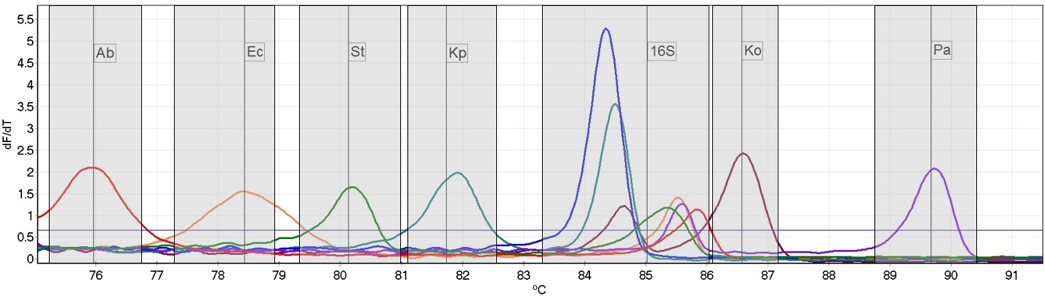
The test can differentiate Klebsiellapneumoniae, Klebsiella oxytoca, Acinetobacter baumannii, Pseudomonasaeruginosa, Escherichia coli and Salmonella enterica, and includes a Gram-negative specific target to identify the presence of other Gram-negative species. The new test was found to be highly accurate when compared with standard PCR and sequencing, with a sensitivity and specificity of 97.1% and 100.0% respectively.
The test uses high resolution melt analysis (HRM) to enable the multiplexing capability necessary to simultaneously test for these 7 targets. This is an endpoint qPCR detection method, that allows discrimination of targets based on amplicon melting temperatures. This technology is also being applied by the diagnostics group to develop tests for markers of drug resistance in both the UK and Malawi, with the goal of enabling improved clinical decision making during bacterial infections.